Here you can find an overview of a few of the research projects I have worked on between 2005 and 2009. For further information or if you have any questions, please do not hesitate to contact me.
Correlative microscopy at liquid nitrogen temperatures
 During the winter semester 2008 and the following summer semester 2009, I was working as a diploma student in the Max Planck Institute of Biochemistry in Martinsried (near Munich) in a department for structural biology. The research of the department focuses on the development of cryo-electron tomography, which is widely seen now as a method of unique potential for visualizing macromolecular complexes in their functional cellular context. It holds great promise for bridging the gap between molecular and cellular structural biology. I was working on correlative microscopy at cryo-temperatures. Correlative microscopy attempts to combine multidimensional information from complementary techniques to bridge the various resolution gaps and thus to be able to integrate structural information gathered from multiple levels of the biological hierarchy into one common framework. I was combining cryo-fluorescence microscopy with cryo-electron tomography. Cryo-fluorescence microscopy is exploited to navigate the cellular landscapes for features of interest before zooming in on these areas by cryo-electron tomography. It offers independent and unambiguous confirmation of the identity of the investigated features. The aim of the project was to optimize and further develop the method. At the end of my diploma thesis entitled “Correlative Microscopy at cryo-temperatures,” I had the opportunity to present my results during a PROSPECTS (PROteomics SPECification in Time and Space) conference.
During the winter semester 2008 and the following summer semester 2009, I was working as a diploma student in the Max Planck Institute of Biochemistry in Martinsried (near Munich) in a department for structural biology. The research of the department focuses on the development of cryo-electron tomography, which is widely seen now as a method of unique potential for visualizing macromolecular complexes in their functional cellular context. It holds great promise for bridging the gap between molecular and cellular structural biology. I was working on correlative microscopy at cryo-temperatures. Correlative microscopy attempts to combine multidimensional information from complementary techniques to bridge the various resolution gaps and thus to be able to integrate structural information gathered from multiple levels of the biological hierarchy into one common framework. I was combining cryo-fluorescence microscopy with cryo-electron tomography. Cryo-fluorescence microscopy is exploited to navigate the cellular landscapes for features of interest before zooming in on these areas by cryo-electron tomography. It offers independent and unambiguous confirmation of the identity of the investigated features. The aim of the project was to optimize and further develop the method. At the end of my diploma thesis entitled “Correlative Microscopy at cryo-temperatures,” I had the opportunity to present my results during a PROSPECTS (PROteomics SPECification in Time and Space) conference.
This project was carried out in the group of Prof Baumeister.
As an introduction to electron microscopy I can highly recommend Transmission Electron Microscopy by Ludwig Reimer and Helmut Kohl and Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State by Joachim Frank.
Rheometer measurements of cytoskeleton and biopolymer networks
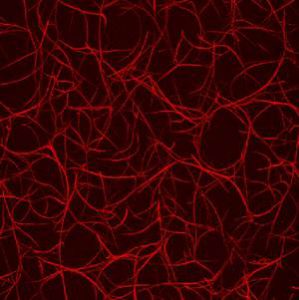 During the summer semester 2008, I was working as a student trainee in a biophysics group at the TU Munich whose research interest is focused on the physics of biological systems, in particular the cytoskeleton. I measured how the presence of certain proteins influences the polymerization rate of actin filaments. To gain information about the rates, I was using a rheometer, a device that measures the response of a solution to applied external forces. Since the viscosity of actin solution increases due to polymerization, the rate was accessible. It was impressive to get to know how outstanding the mechanical properties of materials in nature can be and how they are controlled.
During the summer semester 2008, I was working as a student trainee in a biophysics group at the TU Munich whose research interest is focused on the physics of biological systems, in particular the cytoskeleton. I measured how the presence of certain proteins influences the polymerization rate of actin filaments. To gain information about the rates, I was using a rheometer, a device that measures the response of a solution to applied external forces. Since the viscosity of actin solution increases due to polymerization, the rate was accessible. It was impressive to get to know how outstanding the mechanical properties of materials in nature can be and how they are controlled.
This project was carried out in the group of Prof Bausch.
If you would like to learn more about polymer physics I highly recommend the following books: Introduction to Polymer Physics by M. Doi and Polymer Physics by M. Rubinstein and Ralph H. Colby.
FLIC measurements of HEK cells
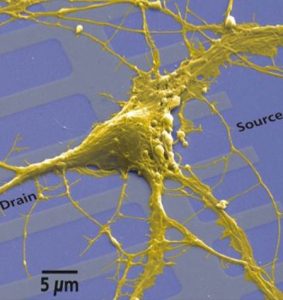 During the spring break in 2008, I did an internship in the Max Planck Institute of Biochemistry in Martinsried (near Munich) in a department for membrane and neurophysics. The department studied the electrical interfacing of semiconductors and of living cells, in particular of neurons. The research was directed to reveal the structure and dynamics of the cell-semiconductor interface and to build hybrid neuroelectronic networks. Therefore cellular processes were coupled to microelectronic devices through direct contact of cell membranes and semiconductor chips. The outcome of such studies will have a huge impact on future applications in medical prosthetics, biosensors, brain research, and neurocomputation. I was analyzing how tightly different types of HEK cells attach to silicon wafers. Fluorescence Interference Contrast (FLIC) Microscopy, which allows the determination of nanometer distances, was used for this purpose. Again it was very impressive to see how biologists, physicists, and engineers work together for a common goal.
During the spring break in 2008, I did an internship in the Max Planck Institute of Biochemistry in Martinsried (near Munich) in a department for membrane and neurophysics. The department studied the electrical interfacing of semiconductors and of living cells, in particular of neurons. The research was directed to reveal the structure and dynamics of the cell-semiconductor interface and to build hybrid neuroelectronic networks. Therefore cellular processes were coupled to microelectronic devices through direct contact of cell membranes and semiconductor chips. The outcome of such studies will have a huge impact on future applications in medical prosthetics, biosensors, brain research, and neurocomputation. I was analyzing how tightly different types of HEK cells attach to silicon wafers. Fluorescence Interference Contrast (FLIC) Microscopy, which allows the determination of nanometer distances, was used for this purpose. Again it was very impressive to see how biologists, physicists, and engineers work together for a common goal.
This project was carried out in the group of Prof Fromherz (emeritus since 2011).
Measuring protein dynamics by neutron scattering
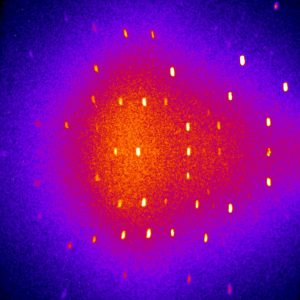 In the summer semester of 2007 and the following winter semester, I was working as a student trainee at a research neutron source. During the first part of my training, I was writing a program to evaluate multi-SANS measurements for the multipurpose instrument MIRA. In the second part, I measured the dynamics of the protein Cytochrom C at the resonance spin echo spectrometer RESEDA.
In the summer semester of 2007 and the following winter semester, I was working as a student trainee at a research neutron source. During the first part of my training, I was writing a program to evaluate multi-SANS measurements for the multipurpose instrument MIRA. In the second part, I measured the dynamics of the protein Cytochrom C at the resonance spin echo spectrometer RESEDA.
This project was carried out in the group of Prof Böni at the research reactor Munich II (FRM II) at the TU Munich.
A nice introduction to neutron scattering by Roger Pynn can be found here.
Furthermore I would like to recommend Introduction to the Theory of Thermal Neutron Scattering by G. L. Squires.
Analysis of DNA with an AFM
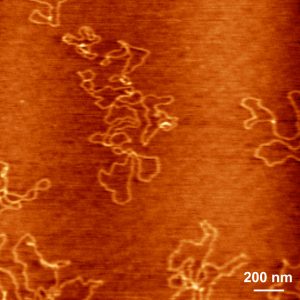 In the summer semester of 2006 and the following winter semester, I was working as a student trainee in a biophysics group at the TU Munich. The research of the group was focused on molecular machines and biopolymer folding and interaction. In my project, I measured with an Atomic Force Microscope how DNA adheres on surfaces as well as how special coatings adhere to plastic.
In the summer semester of 2006 and the following winter semester, I was working as a student trainee in a biophysics group at the TU Munich. The research of the group was focused on molecular machines and biopolymer folding and interaction. In my project, I measured with an Atomic Force Microscope how DNA adheres on surfaces as well as how special coatings adhere to plastic.
This project was carried out in the group of Prof Hugel.
If you would like to learn more about biological physics I highly recommend the following books: Biological Physics by Philip Nelson, Physical Biology of the Cell by Rob Phillips, Mechanics of the Cell by David Boal, Theoretical Molecular Biophysics by Philipp O. J. Scherer and Sighart F. Fischer, Molecular Driving Forces: Statistical Thermodynamics in Biology, Chemistry, Physics, and Nanoscience by Ken Dill and Sarina Bromberg, Intermolecular and Surface Forces by Jacob N. Israelachvili and Principles of Colloid and Surface Chemistry by Paul C. Hiemenz and Raj Rajagopalan.
Performance evaluation of an animal PET scanner
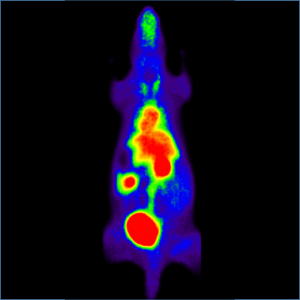 During the spring break in 2006, I did an internship in a hospital in Munich where the latest generation of Positron Emission Tomography (PET) scanners was used. It is a very powerful technique in nuclear medicine to image functional processes in the body. It is used heavily in clinical oncology and for the clinical diagnosis of certain diffuse brain diseases such as those causing various types of dementias. PET is also an important research tool to map the normal human brain and heart function. I was helping scientists to evaluate the performance of an animal PET scanner. Further, I had the possibility to join a one-week hands-on training in which the PET technology was explained to other scientists. It was a thrilling experience to see how researchers, physicians, and engineers work hand in hand.
During the spring break in 2006, I did an internship in a hospital in Munich where the latest generation of Positron Emission Tomography (PET) scanners was used. It is a very powerful technique in nuclear medicine to image functional processes in the body. It is used heavily in clinical oncology and for the clinical diagnosis of certain diffuse brain diseases such as those causing various types of dementias. PET is also an important research tool to map the normal human brain and heart function. I was helping scientists to evaluate the performance of an animal PET scanner. Further, I had the possibility to join a one-week hands-on training in which the PET technology was explained to other scientists. It was a thrilling experience to see how researchers, physicians, and engineers work hand in hand.
This project was carried out in the department of nuclear medicine at the Klinikum rechts der Isar in Munich.
Data analysis of water levels of the Baltic Sea
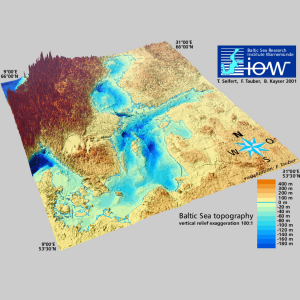 During the fall break in 2005, I did an internship at the Leibniz Institute for Baltic Sea Research, Warnemünde, in the Department of Physical Oceanography and Instrumentation. The department’s main research focus is the investigation and the modeling of the dynamics of marine environments/ecosystems with a focus on the Baltic Sea. To visualize and analyze data, we used MATLAB and Ferret, an interactive computer visualization and analysis environment designed to meet the needs of oceanographers and meteorologists analyzing large and complex gridded data sets. In my project, I analyzed long-term water level data sets of various locations at the Baltic Sea. It was the first time that I worked in a research institution with scientists, and I enjoyed the friendly and inspiring atmosphere at lot.
During the fall break in 2005, I did an internship at the Leibniz Institute for Baltic Sea Research, Warnemünde, in the Department of Physical Oceanography and Instrumentation. The department’s main research focus is the investigation and the modeling of the dynamics of marine environments/ecosystems with a focus on the Baltic Sea. To visualize and analyze data, we used MATLAB and Ferret, an interactive computer visualization and analysis environment designed to meet the needs of oceanographers and meteorologists analyzing large and complex gridded data sets. In my project, I analyzed long-term water level data sets of various locations at the Baltic Sea. It was the first time that I worked in a research institution with scientists, and I enjoyed the friendly and inspiring atmosphere at lot.
This project was carried out in the subgroup “Baltic Sea system dynamics” with Dr Torsten Seifert.
For further information or if you have any questions, please do not hesitate to contact me.